eDNA: Finding Tiny Clues with Big Meaning for Salmon in the Gulf of Alaska
Camille Jasinski, IYS Communications Coordinator
April 26, 2021
Dr. Christoph Deeg saw his first wild salmon in 2006 while he was mountaineering on the Kamchatka Peninsula in Russia, where he says the Chinook salmon spawning in streams were exceptionally huge. In the Kamchatka region, salmon is an integral part of the culture. The locals eat salmon every day – that includes salmon eggs, or salmon roe, every morning for breakfast. Christoph grew up in Germany, and spent time on the North Atlantic and North Seas as a research assistant during his undergraduate degree at Albert-Ludwigs University in Freiburg, where he was studying virology, immunology, molecular genetics, zoology, and ecology. Because of his interest in microbiology, he eventually earned his MSc in virology and went on to complete his PhD in environmental microbiology at the University of British Columbia (UBC). As a PhD candidate at UBC, Christoph went fishing in the Salish Sea, which is where he first came to appreciate the role of salmon as a keystone species in the Pacific Northwest. After realizing how important salmon are to our ecosystems, he dedicated his work to protecting them.

In the winter of 2019, twenty scientists from Canada, Japan, Korea, Russia, and the United States – the five member countries of the North Pacific Anadromous Fish Commission (NPAFC) – embarked on a month-long survey to study the winter ecology of Pacific salmon in the Gulf of Alaska. The expedition, which was conducted on board the Russian R/V Professor Kaganovskiy , aimed to study the mechanisms affecting salmon survival and production in the region. Christoph, who had just finished his PhD at UBC, was one of the scientists on the expedition and succeeded in performing genetic stock identification on board the ship. This was the first time a scientists had successfully conducted this task at sea!
In 2020, Dr. Richard Beamish (DFO Emeritus) and Dr. Brian Riddell (Pacific Salmon Foundation) organized a second expedition to go back into the Gulf of Alaska and build on the survey research from 2019. This time, the expedition took place on the Canadian fishing vessel turned research vessel, the Pacific Legacy No.1 and included 12 scientists from Canada, Russia and the United States, including Christoph. During both expeditions, Christoph took water samples at every sampling station and filtered them for DNA. The scientists were surprised that few salmon predators were observed visually or found in the nets during the expedition. This generated interest in seeing if the environmental DNA (eDNA) could provide clarity on all the species present in the Gulf of Alaska.
Christoph conducts his research at the Pacific Biological Station (PBS) in Nanaimo with Dr. Kristi Miller, who is one of the world’s foremost molecular genetic scientists and head of the Applied Diagnostics, Genomics and Technology Division at PBS. Christoph’s postdoctoral research focuses on eDNA, salmon health (using genomic health assessments), and at-sea stock identification. Together, Christoph and Kristi have been processing samples from the Gulf of Alaska Expeditions – particularly genomic health assessment and eDNA samples.
What is eDNA?
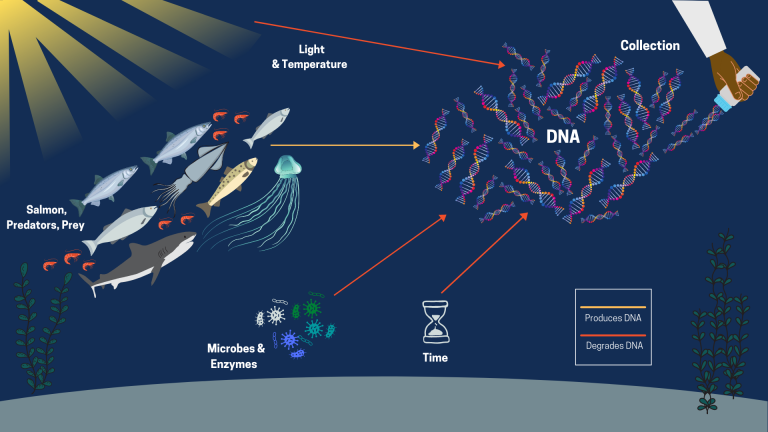
eDNA is a cutting-edge technology in which DNA is collected from environmental samples such as water, soil, or snow. DNA consists of 4 base compounds (represented by the letters: A, C, T, G). These bases can only bind to one other specific base (A to T, C to G), and each organism has a unique sequence of letters that make up their DNA.
Most organisms leave invisible traces of their DNA in the environment, which is derived from things like feces, the sluffing of skin cells/scales, fish milt and eggs, and tissue after injury or death. During the 2019 & 2020 Expeditions, Christoph collected eDNA samples by filtering these cells from water, which includes cellular debris containing DNA.
Once in the lab, the DNA is extracted from the debris and scientists begin to work on amplifying regions of interest. For the Gulf of Alaska Expeditions, these regions were genes which allow for differentiation between salmon and squid species, as well as a gene that is present in all vertebrates, for the detection of salmon predators and other fish. After detection, the amplified DNA was sequenced and compared to a global database of all sequenced genomes to identify what organism they belonged to. Potential predator species detected via eDNA from the 2019 & 2020 Expeditions were beaked whales, sea lions, seals, lancetfish, daggertooth sharks, sperm whales, and baleen whales – a whole community of animals otherwise unseen! eDNA also confirmed the presence of dogfish sharks that were caught in the trawls and detected numerous instances of Dall’s porpoise and killer whales that had been visually observed. The samples also found the presence of pink salmon in the North in 2019, as well as the eastward movement of coho and pink salmon in 2020. Most importantly, Christoph also detected salmon sharks in several samples, which are thought to be the most important predator of salmon in the open ocean.

There are some incredible benefits of eDNA, particularly in a rapidly changing and volatile environment. First of all, there is no need to capture and handle animals in the process. There is also no capture bias in eDNA – it can detect animals that are hard to capture or observe, otherwise known as secretive or evasive species, such as the fast-moving predators of salmon. Generally, collecting eDNA can be relatively cheap since it can be collected on a dinghy or a remotely operated vessel and there is not necessarily the need for a fishing vessel, as people use unmanned drones and gliders to collect eDNA in the high seas. However, as with all scientific methods, eDNA is not without its problems. It can be time consuming, and although collecting eDNA is relatively inexpensive, the genomic sequencing is still very expensive. As a relatively new practice, collecting, sequencing, and analyzing eDNA can get very complicated, and the results themselves are only semi-quantitative, meaning that while the identity of organisms can be determined, it is difficult to ultimately estimate their abundance. There are also risks associated with contamination, and DNA contamination is common. Christoph warns that even eating sushi before sampling for salmon DNA can potentially lead to sample contamination, and jokes that he could recreate the board menu based on eDNA reads!

Plans for eDNA for the 2022 IYS Pan-Pacific Winter High Seas Expedition
With the collaboration of all five NPAFC member countries, the 2022 IYS Pan-Pacific Winter High Seas Expedition will further expand our understanding of salmon and the North Pacific ecosystems. Building on research from 2019 and 2020, the 2022 Expedition aims to advance oceanographic research and global scientific governance, inform efforts to sustain global biodiversity and food security, and use salmon data and research to inform key ocean and climate science. Christoph intends to continue his eDNA research on the 2022 Expedition by conducting a pan-Pacific eDNA survey. This will include building a semi-automated filtration system, which he says will be easy to use for non-specialists.
The legacy of eDNA for the International Year of the Salmon after all three expeditions in the North Pacific Ocean will be building a pan-Pacific baseline of biodiversity. This will help current and future scientists answer questions about the distribution of key organisms in the dynamic environment in which salmon exist. Scientists discussed the results of the 2019 and 2020 Gulf of Alaska Expeditions at the 2021 Conference on the Winter Ecology of Pacific Salmon, which took place from April 20-22, 2021.

